3.2 Designed experiments. Example 3.1
3.3 Mixed models with emphasis on random effects. Example 3.2
3.4 Covariance structures in the mixed model. Example 3.3
The mixed model methodology has been developed within the discipline of animal genetics and breeding (Henderson, 1990). From there, it has spread to many other disciplines, and mixed models have been fitted and used for analysis of various types of data. Many applications in veterinary science are based on mixed rather than fixed effects models.
In this chapter three diverse situations will be described where the appropriate statistical analysis is based on mixed models techniques.
The first field, discussed in Section 3.2, is that of designed experiments, such as randomised complete block, split-plot and latin square designs. In these experiments block parameters may be best described by variance components which describe the background variability against which different hypotheses are tested.
The second field, discussed in Section 3.3, is that of experiments for which there is primary interest in both random and fixed effects. Experiments in this field include breeding experiments (Gianola and Hammond, 1990) and epidemiological studies.
The third field, discussed in Section 3.4, is that of experiments in which observations are correlated with each other, either because they are repeated measurements in time (Diggle et al., 1994) or because they are spatially correlated (Cressie, 1991; Ripley, I981). For such experiments interest is in the fixed effects as well as in the types of correlations among observations.
An experiment can of course consist of a combination of these three categories. For instance, a designed experiment might also have repeated measures, for which the correlation structure must be determined. The three categories will, however, be explained separately in order to focus on the important concepts.
The data are given in Appendix A and the full SAS programs in Appendix B. Relevant information extracted from the SAS output of the programs is shown in this chapter.
To demonstrate how the mixed model can be used for designed experiments, the following experiment is analysed by the mixed model.
Two drugs against trypanosomosis, Berenil and Samorin, are studied in a situation where there is widespread evidence of high levels of drug resistance. Four herds of cattle are randomly assigned to Berenil and four herds to Samorin treatment. It was considered impracticable to organise the study so that both drugs were used in the same herd. Only one dose of Samorin (comparable with the low dose of Berenil) is used. Animals belonging to a herd to which Berenil has been assigned, however, are randomly assigned to receive a high or a low dose of Berenil when detected parasitaemic with trypanosomes.
Packed cell volume was monitored monthly. That determined at the time of treatment was designated PCVl, that a month later following treatment was designated PCV2. A significant increase in PCV reflects a beneficial effect of treatment, despite presence of drug resistance. The data are shown in Appendix A, Example 3.1.
Different models can be fitted to this data structure and three different models with different degrees of complexity are fitted here.
Model 1
In the first model the data structure can be described by
Yijkl = µ + Ti + hij + δik + eijkl
where, for i = 1, 2; j = 1, 2, 3, 4; k = 1, 2 if i = 1; k = 1 if i = 2; l = 1, ..., nijk,
Yijkl is the PCV one month after drug administration (=PCV2)
µ is the overall mean
Ti is the effect of Berenil (Tl) or Samorin (T2)
hij is the effect of herd j to which drug i is assigned
δik is the effect of the low (δ11) or high (δ12) dose of Berenil or the low (δ21) dose of Samorin
eijkl is the random error of animal l with dose k of drug i in herd ij
Each of µ, Ti and δik are fixed effects; hij and eijkl are random effects.
The assumptions are:
the hij are iid N(0, σ![]() )
)
the eijkl are iid N(0,σ 2)
hij and eijkl are independent
In this model, the information on PCV at the time of treatment is not used. The SAS-program that fits this model is shown in Appendix B.3.1.1. The REML-estimates of the variance components can be obtained from the 'Covariance Parameter Estimates (REML)' table:
Covariance Parameter Estimates (REML) | |
Cov Parm |
Estimate |
HERD |
0.2572 |
Residual |
15.4001 |
The variance component describing the variability among animals in the same herd is much larger than that among herds. Thus in this example there is little additional variability among animals from different herds compared with animals from the same herd.
Once estimates of the variance components have been obtained, they can be used to derive estimates of the fixed effects. These estimates can be obtained from the 'Solution for Fixed Effects' table.
Solution for Fixed Effects | |||||||
Effect |
DRUG |
DOSE |
Estimate |
Std Error |
DF |
t |
Pr > ½t½ |
INTERCEPT |
23.891 |
0.442 |
7.28 |
54.03 |
0.0001 | ||
DRUG |
BERENIL |
0.0658 |
0.638 |
8.4 |
0.1 |
0.92 | |
DRUG |
SAMORIN |
0.0000 |
|||||
DOSE (DRUG) |
BERENIL |
1 |
–0.644 |
0.528 |
348 |
–1.22 |
0.224 |
DOSE (DRUG) |
BERENIL |
2 |
0.0000 |
||||
DOSE (DRUG) |
SAMORIN |
1 |
0.0000 |
||||
The model, as described above, is overparameterised, and a solution is thus obtained by ensuring that functions of parameters are estimable. SAS does this by setting some parameter levels to 0. In the solution supplied by SAS, the following parameters are set to 0.
T2 = 0; δ12 = 0; δ21 = 0
Therefore, INTERCEPT in the table is the expected value of PCV2 for any animal l in any herd j treated with the one dose of Samorin and is given by
E(PCV22j1l) = 23.89
The expected values of PCV2 for the three treatments are shown in the 'Least Squares Means' table:
Least Squares Means | |||||||
Effect |
DRUG |
DOSE |
LSMEAN |
Std Error |
DF |
t |
Pr > ½t½ |
DOSE(DRUG) |
BERENIL |
1 |
23.31 |
0.45 |
8.24 |
51.84 |
0.0001 |
DOSE(DRUG) |
BERENIL |
2 |
23.96 |
0.46 |
9.69 |
52.13 |
0.0001 |
DOSE(DRUG) |
SAMORIN |
1 |
23.89 |
0.44 |
7.28 |
54.03 |
0.0001 |
Note that the value 23.89 in the third line is the same as the INTERCEPT described above. The classical hypotheses tests for the fixed effects can be obtained from the 'Tests of Fixed Effects' table. These are based on the F-distribution and the denominator degrees of freedom (DDF) of the F-statistic is obtained by the Satterthwaite procedure.
Taste of Fixed Effects | ||||
Source |
NDF |
DDF |
Type III F |
Pr > F |
DRUG |
1 |
5.56 |
0.2 |
0.674 |
DOSE(DRUG) |
1 |
348 |
1.49 |
0.224 |
NDF refers to the degrees of freedom of the numerator of the F-test; type III F means that the hypothesis being tested is that corrected for the other effects in the model. The first line, DRUG, tests whether there is a difference in the response to the two drugs; the obtained F-value is not significant. The second test compares the two doses of Berenil, and again no significant difference is found. Note the large difference in DDF, denominator degrees of freedom, for the two tests. This is because the alternative drugs are assigned to the herds as a whole, and therefore the comparison between drugs is based on differences between herds, of which there are only eight in the study. On the other hand, the comparison of the two doses of Berenil is based on animals within herds.
Often our interest is in more specific hypotheses than the general ones just described above. Any linear hypothesis can be tested by specifying the hypothesis as a linear combination cTβ = 0 or set of linear combinations CTβ = 0.
We might want to test whether a similar dose of Berenil and Samorin leads to the same response in PCV. This hypothesis is not included in the 'Tests of Fixed Effects' table. The hypothesis of interest can be written as:
H0 : µ11 = µ21 versus Ha : µ11≠ µ21
The null hypothesis can be rewritten as the following linear combination
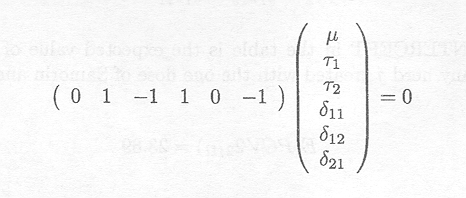
An estimate of this linear combination can be obtained from the third line in the 'ESTIMATE Statement Results' table.
ESTIMATE Statement Results | |||||
Parameter |
Estimate |
Std Error |
DIP |
t |
Pr > ½t½ |
Mann Samorin |
23.89 |
0.442 |
7.28 |
54.03 |
0.0001 |
Berenil dif 2 doses |
– 0.644 |
0.628 |
348 |
–1.22 |
0.224 |
Bar va Sam at dose 1 |
–0.578 |
0.831 |
7.75 |
– 0.92 |
0.387 |
As expected, there is no significant difference between Berenil and Samorin when given in similar doses. The estimated difference is only . 0.578, which is of the same level of magnitude as the standard error which equals 0.631.
The hypotheses present in the 'Tests of Fixed Effects' table can also be tested by rewriting them in terms of appropriate contrasts. For instance, testing whether the two doses of Berenil lead to the same response can be done by testing the hypothesis

and this leads, as shown in the second line of the 'CONTRAST Statement Results' table, to the same test as the one described in the 'Tests of Fixed Effects' table.
CONTRAST Statement Results | ||||
Source |
NDF |
DDF |
F |
Pr > F |
Mean Samorin |
1 |
7.28 |
2919.6 |
0.0001 |
Berenil dif 2 doses |
1 |
348 |
1.49 |
0.224 |
Ber vs Sam at dose 1 |
1 |
7.75 |
0.84 |
0.387 |
Some difference |
2 |
21.1 |
0.85 |
0.52 |
Furthermore, means can be estimated by specifying the appropriate linear combination. For instance, the mean PCV for Samorin can be obtained by estimating

which leads to the same results as shown before in the 'Least Squares Means' table.
Finally, more complex hypotheses can be tested, consisting of different independent linear combinations.
If we want to test whether there exists any genuine difference between the three treatments, expressed as
H0 : µ11 =µ12 = µ21 versus Ha : µik s are not equal
it can be translated as
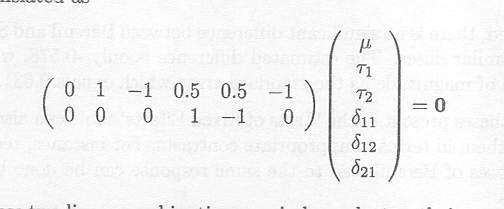
Note that these two linear combinations are independent as their vector product equals 0. The first combination tests whether there is a difference between the two drugs, averaging the response for the high and the low dose for Berenil, whereas the second linear combination tests whether there is a difference between the low and the high dose of Berenil. As we have three treatment means, testing any two independent linear combinations simultaneously corresponds to testing the above hypothesis.
The actual F-value for this hypothesis testing problem is shown in the last line of the 'CONTRAST Statement Results' table, and again, as expected, the null hypothesis is not rejected.
Model 2
Information on the PCV of the animals at the time of treatment is not used in the above analysis. The analysis might be made more powerful if this is used. We indeed expect that animals with very low PCV at the time of treatment may benefit more from the administration of the drug and demonstrate a higher increase in PCV. In order to analyse this type of response the following model is fitted
Yijkl = µ +β x (PCV1ijkl) + Ti + hij + δik + eijkl
where, for i = 1, 2; j = 1, 2, 3, 4; k = 1, 2 if i = l; k = 1 if i = 2; l = 1, ..., nijk,
β is the slope that describes the linear trend between PCV at time of treatment PCV1ijkl and PCV one month later
all other parameters having the same meaning as in Model 1.
The SAS program that fits this model is shown in Appendix B.3.L2.
Estimates for the parameters can be obtained again from the 'Solution for Fixed Effects' table.
Solution for Fixed Effects | |||||||
Effect |
DRUG |
DOSE |
Estimate |
Std Error |
DF |
t |
Pr> ½t½ |
INTERCEPT |
18.548 |
1.935 |
306 |
9.59 |
0.0001 | ||
PCVl |
0.286 |
0.094 |
347 |
2.82 |
0.0051 | ||
DRUG |
BERENIL |
0.249 |
0.806 |
9.4 |
0.41 |
0.69 | |
DRUG |
SAMORIN |
0.0000 |
|||||
DOSE(DRUG) |
BERENIL |
1 |
–0.887 |
0.523 |
346 |
–1.31 |
0.19 |
DOSE(DRUG) |
BERENIL |
2 |
0.0000 |
||||
DOSE(DRUG) |
SAMORIN |
1 |
0.0000 |
||||
It follows from this table that there is indeed a significant effect of the PCV measured at the time of treatment on the PCV one month after treatment. The effect of initial PCV on the response variable can be separately determined for each treatment group. For instance, for the animals treated with the low dose of Berenil, this relationship is given by
E(Y1j1l½PCV11j1l = x ) = 18.548 + 0.266x + 0.249 - 0.687
= 18.11 + 0.266x
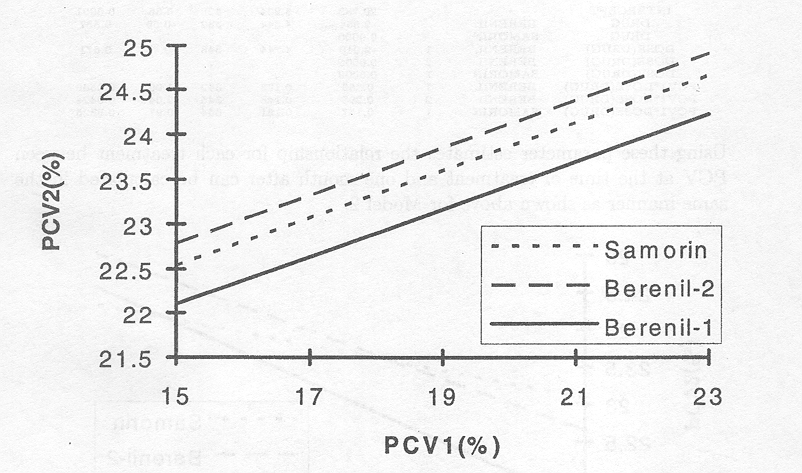
Figure 3.2.1 The effect of initial PCV on PCV one month after treatment for the three treatments based on Model 2.
Although Figure 3.2.1 indicates that the regression line for the lower dose of Berenil falls below the other two, the 'Solution for Fixed Effects' table above indicates no significant mean differences between treatments. Thus, whereas the increase in PCV is greater for lower PCV1, responses were overall similar for each treatment.
Model 3
In Model 2, three regression lines with the same slope were fitted for the three treatments to relate PCV2 to PCVl. Now we relax this assumption and fit a model that allows a different slope for each treatment group. This leads to the following model
Yjkl = µ + βik x (PCV1ijkl ) + Ti + hij + δik + eijkl
where, for i =1, 2; j = 1, 2, 3, 4; k = 1, 2 if i = l; k = 1 if i = 2; l = 1, ..., nijk,
βik is the fixed effect of PCVl at time of treatment on PCV one month later for dose k of drug i
all other parameters having the same meaning as in Model 1
The SAS program that fits this model is shown in Appendix B.3.1.3. Estimates for the parameters can be obtained again from the 'Solution for Fixed Effects' table.
Solution for Fixed Effects | |||||||
Effect |
DRUG |
DOSE |
Estimate |
Std Error |
DF |
t |
Pr > ½t½ |
INTERCEPT |
20.733 |
3.264 |
328 |
6.35 |
0.0001 | ||
DRUG |
BERENIL |
– 2.554 |
4.344 |
332 |
– 0.59 |
0.557 | |
DRUG |
SAMORIN |
0.0000 |
|||||
DOSE(DRUG) |
BERENIL |
I |
– 2.019 |
4.744 |
343 |
– 0.43 |
0.671 |
DOSE(DRUG) |
BERENIL |
2 |
0.0000 |
||||
DOSE(DRUG) |
SAMORIN |
I |
0.0000 |
||||
PCVl*DOSE(DRUG) |
BERENIL |
1 |
0.366 |
0.193 |
342 |
1.90 |
0.0588 |
PCVl*D03E(DRUG) |
BERENIL |
2 |
0.298 |
0.146 |
345 |
2.04 |
0.0424 |
PCVl*DOSE(DRUG) |
SAMORIN |
1 |
0.157 |
0.161 |
344 |
0.97 |
0.3325 |
Using these parameter estimates the relationship for each treatment between PCV at the time of treatment and one month after can be calculated in the same manner as shown above for Model 2.
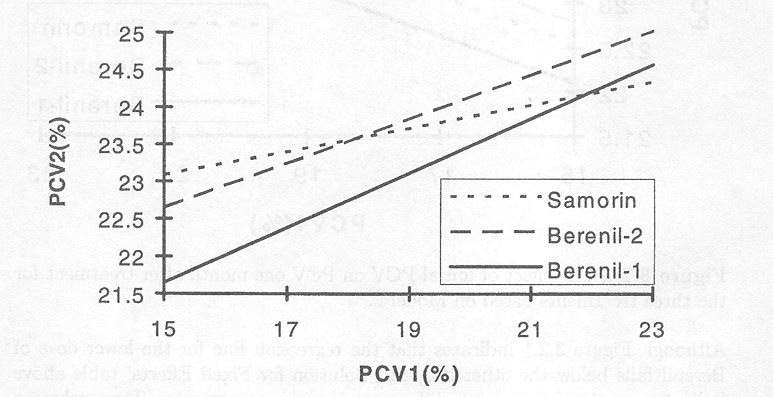
Figure 3.2.2 The effect of initial PCV on PCV one month after treatment for the three treatments based on Model 3.
The figure shows that, whichever treatment is used, the higher the PCV at the time of treatment, the lower its increase in response to treatment. On the other hand, when PCVl was low both Samorin and Berenil at the high dose appeared to result in higher increases in PCV1 than Berenil at the low dose. The 'Solution for Fixed Effects' table demonstrates that the slope for Samorin is not significant.
An investigator might be interested in estimating how an animal might benefit from the treatment for different values of PCV. This might lead to a recommendation on the value to which PCV needs to fall before treatment should be administered. For a value of PCVl=x, the expected response in PCV for drug i at dose k is given by
E(Yjkl /PCV1ijkl = x) = µ + βiikx + Ti + δik
An estimate for PCV for animals with PCVl=18 % at time of treatment with the low dose of Berenil, for instance, is given by:
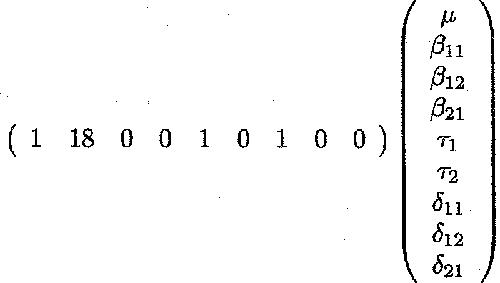
Estimates for PCV1=18 can be obtained from the 'Least Squares Means' table.
Least Squares Means | ||||||||
Effect |
DRUG |
DOSE |
PCV |
LSMEAN |
Std Error |
DF |
t |
Pr > | t | |
DOSE(DRUG) |
BERENIL |
1 |
18 |
22.74 |
0.52 |
20.1 |
43.77 |
0.0001 |
DOSE(DRUG) |
BERENIL |
2 |
IS |
23.54 |
0.48 |
15.2 |
48.98 |
0.0001 |
DOSE(DRUG) |
SAMORIN |
1 |
18 |
23.55 |
0.55 |
17.2 |
44.42 |
0.0001 |
A 95% confidence interval for the mean PCV of animals treated with the low dose of Berenil is then given by
22.74 ± t0.025; 20 x 0.52 = 22.74 ± 1.08
Thus an effective increase in PCV of 4.74 ± 1.08% would have occurred as a result of treating animals with a PCV of 18%.
The emphasis in breeding experiments is on the variance components and on the prediction of particular random effects, but estimation of fixed effects and hypothesis testing with regard to fixed effects is also important. In epidemiological studies both random and fixed effects may be of interest.
An animal breeding experiment is used to illustrate this. A portion of the data set is shown in Appendix A, Example 3.2. The goal of the research is to select for improved helminth resistance in sheep. The female sheep used in the experiment are from three different breeds, whereas the males are from two breeds. In each of the six crosses, there are at least 25 and at most 42 different sires, and each sire within a cross breed has on average offspring of 6.4 lambs. The weaning weight is measured for each of the lambs. The age at which lambs are weaned (AGEW) may differ from animal to animal, and therefore, a variable expressing the age of the animal at weaning is included as a fixed effect in the model. Also the sex (SEX) of the lamb is added to the model as a fixed effect. Finally, the sire is included as a random effect: Although the same sire is mated to ewes from different breeds, the sire nested in breed is taken as a single random effect and it is assumed that these random effects are independent.
An appropriate model for this data structure is thus given by
Yijkl = µ + yi + sij + Tk + β (AGEW) + eijl
where, for i = 1, ..., 6; j = 1, ..., ni; k = 1, 2; l = 1, ..., nij;
Yijkl is the weaning weight of lamb l of sex k from sire j within breed i
µ is the overall mean
yi is the effect of breed i
sij is the effect of sire j within breed i
Tk 1S the effect of sex: female (T1) or male (T2)
β is the effect of age at weaning on weaning weight
eijl is the random error of lamb l from sire j within breed i
Each of µ,β , yi and Tk are fixed effects, whereas sij and eijl are random effects with the following assumptions:
the sij are iid N(0, σ![]() )
)
the eijl are iid N(0,σ 2)
sij and eijl are independent
The SAS-program that fits this model is given in Appendix B.3.2. The REML-estimates of the variance components can be obtained from the 'Covariance Parameter Estimates (REML)' table.
Covariance Parameter Estimates (REML) | |
Cov Parm |
Estimate |
SIRE-ID(BREED) |
0.8802 |
Residual |
4.8639 |
A commonly derived statistic is the heritability (h2) of the trait, which is a function of these two variance components and describes the proportion of the total variation in the environment of this study attributable to inheritance from the lambs' parents.
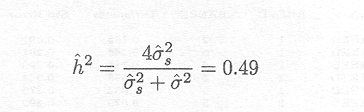
In this formula ![]()
![]() is multiplied by 4 in the numerator to account for the fact that lambs from the same sire are half siblings and the sire accounts for half of the inherited genetic component, and (
is multiplied by 4 in the numerator to account for the fact that lambs from the same sire are half siblings and the sire accounts for half of the inherited genetic component, and (![]()
![]() +
+ ![]() 2) is the phenotypic variance. The higher the heritability, which lies between 0 and 1, the higher the proportion of total variation that can be assumed to be genetic.
2) is the phenotypic variance. The higher the heritability, which lies between 0 and 1, the higher the proportion of total variation that can be assumed to be genetic.
Once estimates of the variance components have been obtained, they can be used to derive estimates of the fixed effects. These estimates can be obtained from the 'Solution for Fixed Effects' table.
Solution for Fixed Effects | |||||||
Effect |
SEX |
BREED |
Estimate |
Std Error |
DF |
t |
Pr > ½t½ |
INTERCEPT |
5.003 |
0.537 |
949 |
9.32 |
0.0001 | ||
SEX |
1 |
– 0.704 |
0.128 |
1229 |
– 5.51 |
0.0001 | |
SEX |
2 |
0.0000 |
|||||
BREED |
1 |
1.682 |
0.376 |
327 |
4.47 |
0.0001 | |
BREED |
2 |
1.515 |
0.383 |
290 |
3.96 |
0.0001 | |
BREED |
3 |
1.436 |
0.357 |
299 |
4.02 |
0.0001 | |
BREED |
4 |
0.948 |
0.357 |
262 |
2.66 |
0.0084 | |
BREED |
5 |
– 0.406 |
0.393 |
274 |
– 1.03 |
0.3025 | |
BREED |
6 |
0.0000 |
|||||
AGEW |
0.0465 |
0.0033 |
1214 |
14.02 |
0.0001 | ||
There is a significant effect of sex with female lambs weighing on average 0.704 kg less at weaning than males. Furthermore, there is a significant effect of age at weaning with weaning weight increasing by 0.046 kg per daily increase in age. There are also significant differences among breeds, with breeds 1– 4 having significantly higher weaning weights than breeds 5– 6. Breed means axe given in the 'Least Squares Means' table.
Least Squares Means | ||||||
Effect |
BREED |
LSMEAN |
Std Error |
DF |
t |
Pr >½t½ |
BREED |
1 |
12.48 |
0.222 |
216 |
56.23 |
0.0001 |
BREED |
2 |
12.31 |
0.232 |
170 |
53.00 |
0.0001 |
BREED |
3 |
12.23 |
0.187 |
145 |
65.21 |
0.0001 |
BREED |
4 |
11.75 |
0.187 |
102 |
62.79 |
0.0001 |
BREED |
5 |
10.39 |
0.249 |
160 |
41.70 |
0.0001 |
BREED |
6 |
10.80 |
0.304 |
420 |
35.54 |
0.0001 |
To compare these means we can use a multiple comparisons procedure, such as Tukey-Kramer (see SAS Institute Inc. Cary, NC, 1996 on the procedure MULTTEST and Westfall and Young, 1993). The results are given in the underlying 'Differences of Least Squares Means' table. Some of the columns of the original table have been deleted.
Differences of Least Squares Means | |||||
Effect |
BREED |
_BREED |
Difference |
Std Error |
Adj. P |
BREED |
1 |
2 |
0.166 |
0.321 |
0.995 |
BREED |
1 |
3 |
0.246 |
0.291 |
0.958 |
BREED |
1 |
4 |
0.733 |
0.290 |
0.122 |
|
BREED |
1 |
5 |
2.088 |
0.334 |
0.000 |
|
BREED |
1 |
6 |
1.682 |
0.376 |
0.000 |
|
BREED |
2 |
3 |
0.079 |
0.299 |
0.999 |
|
BREED |
2 |
4 |
0.567 |
0.298 |
0.406 |
|
BREED |
2 |
5 |
1.921 |
0.342 |
0.000 |
|
BREED |
2 |
6 |
1.515 |
0.382 |
0.001 |
|
BREED |
3 |
4 |
0.487 |
0.265 |
0.443 |
|
BREED |
3 |
5 |
1.842 |
0.312 |
0.000 |
|
BREED |
3 |
6 |
1.436 |
0.357 |
0.001 |
|
BREED |
4 |
5 |
1.354 |
0.312 |
0.000 |
|
BREED |
4 |
6 |
0.948 |
0.357 |
0.089 |
|
BREED |
5 |
6 |
–0.406 |
0.393 |
0.906 |
From this table, it can be concluded that significant differences exist when comparing breed 1, 2, 3 and 4 with breed 5 and 6, but not among 1, 2, 3 and 4 nor among 5 and 6.
Apart from estimating heritability and comparing breed means, an investigator might be interested in the breeding value of specific sires in order to determine which sires should be retained for future selection. As seen before, such a prediction of the breeding value of a sire can be obtained by best linear unbiased prediction. For each sire, such a prediction can be obtained from the 'Solution for Random Effects' table.
|
Effect |
SIRE-ID |
BREED |
Estimate |
SE Pred |
DF |
t |
Pr >½t½ |
|
SIRE_ID(BREED) |
1971 |
1 |
–0.1061 |
0.6653 |
85.9 |
–0.16 |
0.874 |
|
SIRE-ID(BREED) |
1972 |
1 |
0.5241 |
0.5349 |
189 |
0.98 |
0.328 |
|
SIRE_ID(BREED) |
1973 |
1 |
0.3888 |
0.6399 |
99.9 |
0.61 |
0.545 |
|
SIRE_ID(BREED) |
1974 |
1 |
1.9339 |
0.5486 |
174 |
3.53 |
0.000 |
The predicted breeding value for sire with ID 1974 equals 1.93, meaning that offspring of this sire are predicted to have a weaning weight that is 1.93 kg higher than the average offspring of all sires of the same breed.
An important assumption in the use of the fixed effects model is that observations are independent from each other. In many practical situations this assumption may not hold. The most common situation is where different measurements are taken on the same individual, leading to what is known as a repeated measures design. Often these measurements are taken periodically over time. Alternatively, observations may be spatially collected in which case those closest together may be most alike.
As an example, we study the repeated measures design introduced in Example 1.2.8.
The aim of the study was to see whether there are differences in the change in PCV between the two breeds following a trypanosome infection. It is clear that observations collected over time on the same animal are correlated. The data set is shown in Appendix A, Example 3.3. Different models can be fitted to the data with different assumed covariance structures.
Model 1
The data structure can be described by the following statistical model
Yikl = µ + βi0 +β iltl +aik + eikl
where, for i = 1, 2; k = 1, ..., 6; l = 1, ...14
Yikl is the PCV of animal k of breed i at time l
µ is the overall mean
βi0 is the intercept of the linear regression line for breed i
βi1 is the slope of the linear regression line for breed i
tl is the time of measurement l
aik is the effect of animal k belonging to breed i
eikl is the random error of measurement l on animal k of breed i
Both aik and eikl are random effects, independent from each other, with the assumptions
the ik are iid N(0, σ![]() )
)
the eikl are iid N(0,σ 2)
From this model structure, it follows that
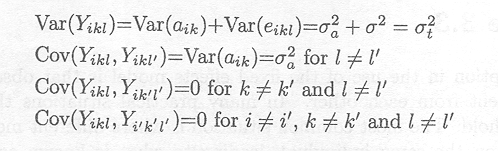
We introduce a new notation, putting the suffix l of Yikl into brackets to clearly separate suffix k from suffix l which can consist of two numbers (l goes from 1 to 14). Thus Yikl = Yik(l).
Therefore, the covariance structure of the 14 observations Yik = (Yik(1), ..., Yik(14)) on the same animal, W = D(Yik), is given by
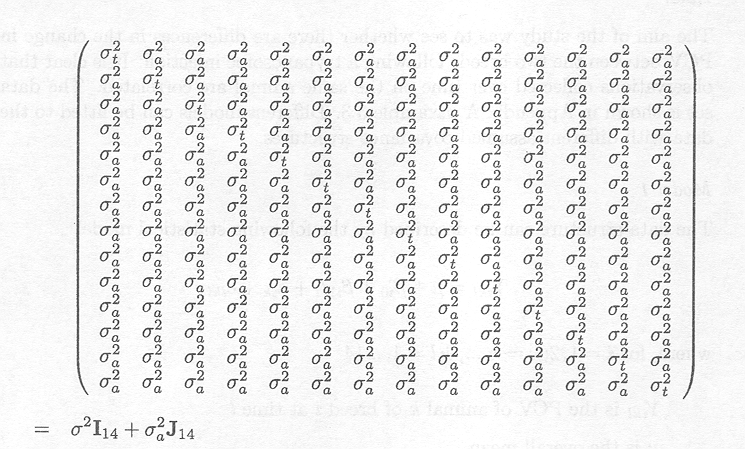
As measurements on different animals are independent from each other, the variance-covariance matrix of the observation vector Y is given by

An alternative model representation can thus be given by
Yikl = µ + βi0 + βiltl + eikl
where we drop the independence assumption on the eikl. The within-subject correlations, modelled by the random effects factor aik; in the first model representation, are now modelled by specifying a more complex variance-covariance matrix for eikl (=eik(l)).
For the random errors of an individual animal, eik =(eik(1), ..., eik(14)), is it now assumed that
eik ~ MVN(0, σ 2I14 + σ
J14
and also that
D(eik) = D(Yik) and D(e) - D(Y) = V
This type of covariance structure is called compound symmetry with each observation on an animal having the same correlation with each other observation measured on the same animal.
The advantage of the first model specification is that it uses the general notation for the mixed model and it shows how the covariance structure can be expressed in terms of two random effects. The disadvantage of the first model specification is that it can only deal with the compound symmetry structure. If a more complex covariance structure is assumed, the second model specification needs to be used, as will be shown in Model 2.
The SAS-program that fits these two model specifications is given in Appendix B.3.3.1. Since the two model specifications lead to the same results, we only discuss here the results from the first model specification.
The REML-estimates of the variance components can be obtained from the 'Covariance Parameter Estimates (REML)' table.
|
Covariance Parameter Estimates (REML) | |
|
Cov Parm |
Estimate |
|
ANIM_ID(BREED) |
5.478 |
|
Residual |
4.965 |
The covariance parameter 'ANIM-ID(BREED)' equivalent to
the variance component σ![]() in the model corresponds to the
covariance among measurements within an animal (see matrix D(Yik) above).
in the model corresponds to the
covariance among measurements within an animal (see matrix D(Yik) above).
Based on these results, we can conclude that the
estimated covariance between two measurements within the same animal is equal to
5.478, whereas the estimated variance of a single measurement (given by σ2+σ![]() ) is 5.478+4.965=10.443.
) is 5.478+4.965=10.443.
Estimates can be obtained from the 'Solution for Fixed Effects' table.
|
Solution for Fixed Effects | ||||||
|
Effect |
BREED |
Estimate |
Std Error |
DF |
t |
Pr > ½t½ |
|
INTERCEPT |
35.06 |
1.056 |
10 |
33.21 |
0.0001 | |
|
BREED |
BO |
– 0.842 |
1.493 |
10 |
– 0.56 |
0.585 |
|
BREED |
ND |
0.00000 |
||||
|
TIME*BREED |
BO |
– 0.413 |
0.0225 |
154 |
– 18.38 |
0.0001 |
|
TIME*BREED |
ND |
– 0.276 |
0.0225 |
154 |
– 12.28 |
0.0001 |
From this table, the linear changes in PCV of the two breeds with time can be obtained as
BO: PCV = 35.06 – 0.842 - 0.413t
ND: PCV = 35.06 - 0.276t
Thus, PCVs of the Borans appear to decrease more rapidly than those of the N'Damas. To determine whether this is statistically significant a contrast can be written
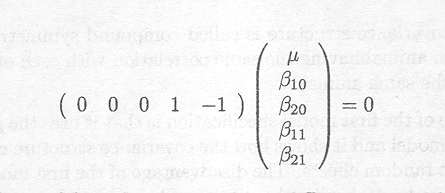
Alternatively, the model can be expressed in a different way with time as fixed effect and the time*breed term now representing the interaction between time and breed. The model can be written as
Yikl = µ + βi0 + β1tl + βi1tl + eikl
where βi1 now represents the difference in slope for breed i from the average.
The program fitting this model is given as the third mixed procedure statement in Appendix B.3.3.1. The significance test for this interaction can be found from the 'Tests of Fixed Effects' table.
Tests of Fixed Effects
|
Source |
NDF |
DDF |
Type III F |
Pr > F |
|
TIME |
1 |
154 |
470.06 |
0.0001 |
|
BREED |
1 |
10 |
0.32 |
0.585 |
|
TIME-BREED |
1 |
154 |
18.62 |
0.0001 |
This table shows that the rate of decrease is different for the two breeds as indicated by the significant interaction term. On the other hand, the intercept of the two lines do not differ significantly as shown by the F-test for breed.
The slopes for the two breeds are shown in the 'Solution for Fixed Effects' table above. These are fixed effects and assumed to be the same for each animal within a breed. However, having introduced a random animal effect into the model, a different intercept can be calculated for each animal. The intercept for a particular animal can be obtained from the 'Solution for Random Effects' table.
|
Solution for Random Effects | |||||||
|
Effect |
BREED |
ANIM-ID |
Estimate |
SE Pred |
DF |
t |
Pr >½ t½ |
|
ANIM-ID(BREED) |
BO |
BO241 |
– 0.672 |
1.091 |
154 |
– 0.62 |
0.539 |
|
ANIM-ID (BREED) |
BO |
BO322 |
– 2.497 |
1.091 |
154 |
– 2.29 |
0.023 |
|
ANIM-ID(BREED) |
BO |
BO326 |
– 2.383 |
1.091 |
154 |
– 2.18 |
0.030 |
|
ANIM-ID(BREED) |
BO |
BO209 |
3.0379 |
1.091 |
154 |
2.78 |
0.006 |
|
ANIM-ID(BREED) |
BO |
BO37 |
0.0324 |
1.091 |
154 |
0.03 |
0.976 |
|
ANIM-ID(BREED) |
BO |
BOl |
2.481 |
1.091 |
154 |
2.27 |
0.024 |
|
ANIM-ID(BREED) |
ND |
ND60 |
– 1.667 |
1.091 |
154 |
– 1.53 |
0.128 |
|
ANIM-ID(BREED) |
ND |
ND66 |
2.626 |
1.091 |
154 |
2.41 |
0.0173 |
|
ANIM-ID(BREED) |
ND |
ND72 |
– 1.667 |
1.091 |
159 |
– 1.53 |
0.128 |
|
ANIM-ID(BREED) |
ND |
ND73 |
– 0.372 |
1.091 |
154 |
– 0.34 |
0.733 |
|
ANIM-ID(BREED) |
ND |
ND74 |
– 1.754 |
1.091 |
154 |
– 1.61 |
0.109 |
|
ANIM-ID(BREED) |
ND |
ND75 |
2.834 |
1.091 |
154 |
2.60 |
0.0103 |
For instance, for the first animal, BO241, the fitted relationship is equal to
BO241 : PCV = 34.22 – 0.672 – 0.413t
PCV = 33.55 – 0.413t
These relationships can be determined for each animal and are presented in the following figure
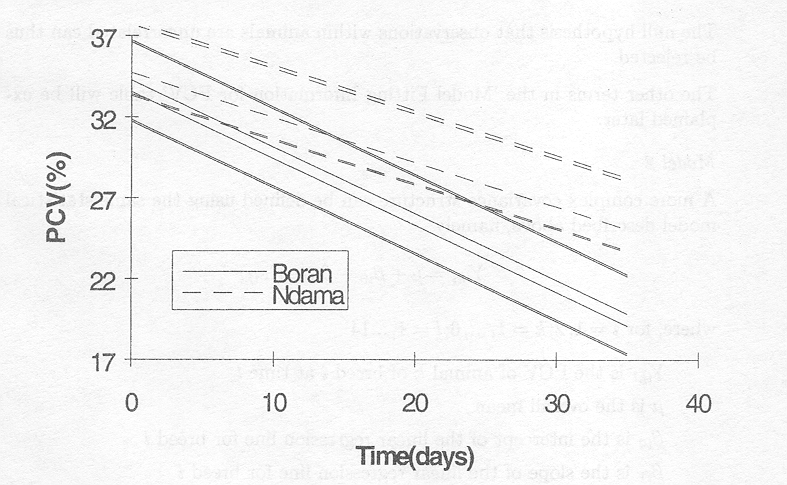
Figure 3.4.1 Change of PCV in time for individual animals based on Model 1. Bold dots and dashes indicate more than one animal with this line.
The second model specification leads to the same results, although the presentation of the results is somewhat different. An interesting additional feature in the output of the second model specification comes from the 'Model Fitting Information for PCV' table.
|
Model Fitting Information for PCV | |
|
Description |
Value |
|
Observations |
168.000 |
|
Res Log Likelihood |
– 391.73 |
|
Akaike's Information Criterion |
– 393.73 |
|
Schwarz's Bayesian Criterion |
– 396.83 |
|
-2 Ras Log Likelihood |
783.451 |
|
Null Modal LRT Chi-Square |
80.848 |
|
Null Model LRT DF |
1.0000 |
|
Null Model LRT P-Value |
0.0000 |
The last three lines contain the information that can be
used to compare the suitability of the model with the compound symmetry
covariance structure with that with the independent covariance structure (Null
Model). As the compound symmetry covariance structure model is nested within the
independent covariance structure model in the sense that the latter structure
contains an additional term σ![]() , the null hypothesis that the two models
describe the data equally well can be tested via the likelihood ratio test
(LRT), which equals 80.845. As the nested model contains one more parameter, the
likelihood ratio statistic is asymptotically Chi-square distributed with one
degree of freedom and the P-value is
, the null hypothesis that the two models
describe the data equally well can be tested via the likelihood ratio test
(LRT), which equals 80.845. As the nested model contains one more parameter, the
likelihood ratio statistic is asymptotically Chi-square distributed with one
degree of freedom and the P-value is
Prob(X![]() ≥ 80.845) <
0.00001
≥ 80.845) <
0.00001
The null hypothesis that observations within animals are uncorrelated can thus be rejected.
The other terms in the 'Model Fitting Information for PCV' table will be explained later.
Model 2
A more complex covariance structure can be defined using the same statistical model described above, namely
Yikl = µ +βi0 +βi1 tl + eikl
where, for i = 1, 2; k = 1, ..., 6; l = 1, ...14
Yikl is the PCV of animal k of breed i at time l
µ is the overall mean
βi0 is the intercept of the linear regression line for breed i
βi1 is the slope of the linear regression line for breed i
tl is the time of measurement l
eikl is the random error of measurement l on animal k of breed i
As before, the random error terms within animals are not assumed independent, but can now be defined to have another and more complex covariance structure. Such a more complex covariance structure D(eik) for the random error term is the first-order autoregressive structure given by

In this matrix, 0 < p < 1 represents the correlation between two observations adjacent in time. This way of modelling the covariance structure means that observations on the same animal that are further apart in time have smaller correlation. The SAS program that fits this model is given in Appendix B.3.3.2. The residual variance σ2 and the correlation p can be obtained from the 'Covariance Parameter Estimates (REML)' table.
|
Covariance Parameter Estimates (REML) | ||
|
Cov Parm |
Subject |
Estimate |
|
AR(1) |
ANIM_ID(BREED) |
0.621 |
|
Residual |
10.191 | |
The estimate for σ2 is 10.191 and the estimate for p is 0.621. The covariance between adjacent observations on the same individual is thus 0.621 x 10.191 = 6.33. The covariance between observations on the same animal decreases the more the observations are apart in time. For instance, the covariance between an observation at time 1 and time 5 or between an observation at time 2 and time 6 equals
Cov(Yik1Yik5) = Cov(Yik2, Yik6) = σ2p4
This covariance is estimated as ![]() 2
2![]() 4= 1.52.
4= 1.52.
T he covariance between an observation at time 1 and time 10 or between an observation at time 3 and time 13 equals
Cov(Yik1, Yik10) = Cov(Yik3, Yik13) = σ2p9
The estimate is 0.141.
To compare Model 1 and Model 2 we cannot use the likelihood ratio test since neither of these models are nested within each other. The models however do have the same number of parameters. We therefore choose a model with the higher value for the log-likelihood. This information can be obtained from the 'Model Fitting Information for PCV' table.
|
Model Fitting Information for PCV | |
|
Description |
Value |
|
Observations |
168.000 |
|
Res Log Likelihood |
–396.57 |
|
Akaike's Information Criterion |
–398.57 |
|
Schwars's Bayesian Criterion |
–401.67 |
|
-2 Res Log Likelihood |
793.139 |
|
Null Model LRT Chi-Square |
71.156 |
|
Null Model LRT DF |
1.0000 |
|
Null Model LRT P-Value |
0.0000 |
The restricted log-likelihood, which is the log-likelihood for an independent set of linear combinations KY as defined in Section 2.4.2, for this model is -396.57. In the corresponding table for the compound symmetry model the restricted log-likelihood equals –391.73. Thus the compound symmetry covariance structure has a slightly higher log-likelihood and thus describes the data better. In a similar way, other covariance structures can be fitted and compared. The two other criteria 'Akaike's Information Criterion' (Akaike, 1974) and 'Schwarz's Bayesian Criterion' (Schwarz, 1978) can also be used to compare models, especially if they contain different number of parameters. They are both based on the log-likelihood, but are penalised for the number of parameters estimated. The larger the value the better the model fits the data. In this case, with equal number of parameters for the two models, these alternative criteria lead to the same conclusion as the restricted log-likelihood.
Model 3
In Models 1 and 2, average slopes for the two breeds are fitted as fixed effects. Thus, the decreases in PCV with time are assumed to be parallel for all animals within a breed, i.e. the slope is the same. This assumption can be further relaxed by assuming that each animal has also a random slope, and that this random slope is normally distributed.
This random coefficients model (Longford, 1993) is given by
Yikl = µ +βi0 + βi1 tl + aik + biktl + eikl
where, for i = 1, 2; k = 1, ..., 6; l = 1, ...14,
Yikl is the PCV of animal k of breed i at time l
µ is the overall mean
βi0 is the intercept of the linear regression line for breed i
βil is the slope of the linear regression line for breed i
aik is the random intercept of animal k of breed i
bik is the random slope of animal k of breed i
tl is the time of measurement l
eikl is the random error of measurement l on animal k of breed i
Both aik, bik and eikl are random effects with the assumptions
the aik are iid N(0, σ![]() )
)
the bik are iid N(0, σ![]() )
)
the eikl are iid N(0,σ 2)
Moreover, we assume that both aik and bik are independent from eikl but that aik and bik are possibly correlated.
The program that fits this model is shown in Appendix B.3.3.3.
The variance components of the random effects factors can be obtained from the 'Covariance Parameter Estimates (REML)' table.
|
Covariance Parameter Estimates (REML) | ||
|
Cov Parm |
Subject |
Estimate |
|
UN(1,1) |
ANIM_ID(BREED) |
12.46457359 |
|
UN(2,1) |
ANIM_ID(BREED) |
–0.2551 |
|
UN(2,2) |
ANIM_ID(BREED) |
0.005758 |
|
Residual |
4.3530421 | |
The first line in this table, named 'UN(l,l)', corresponds to the variability of the intercepts within breed, the second line, named 'UN(2,1)', to the covariance between the intercept and the slope, and the third line, named 'UN(2,2)', to the variability of the slopes within breed. The variance of the intercepts is the largest component and much larger than the variance of the slopes. There is a negative covariance of –0.2551 between the slope and the intercepts, implying that the higher the initial PCV the greater its subsequent fall.
The intercept and slope parameters describing the linear
relationship between PCV contain both fixed and random effects. Average fixed
intercepts and slopes are assumed for each breed, and each animal has a random
slope and intercept, normally distributed with mean 0 and variances σ![]() and σ
and σ![]() respectively.
respectively.
The average linear relationship for each breed can be obtained from the 'Solution for Fixed Effects' table.
|
Solution for Fixed Effect | ||||||
|
Effect |
BREED |
Estimate |
Std Error |
DF |
t |
Pr > ½t½ |
|
INTERCEPT |
35.06 |
1.501 |
10 |
23.35 |
0.0001 | |
|
BREED |
BO |
– 0.842 |
2.123 |
144 |
– 0.4 |
0.692 |
|
BREED |
ND |
0.00000 |
||||
|
TIME*BREED |
BO |
– 0.413 |
0.0374 |
144 |
– 11.04 |
0.0001 |
|
TIME*BREED |
ND |
– 0.276 |
0.0375 |
144 |
– 7.37 |
0.0001 |
From this table, the linear regression equation for the two breeds can be obtained as
BO: PCV = 34.22 – 0.413t
ND: PCV = 35.06 – 0.276t
which is the same as that of Model 1. However, both the intercept and the slope of the fitted relationship is now different from animal to animal.
Individual regression lines for animals can be obtained from the 'Solution for Random Effects' table.
|
Solution for Random Effects | ||||||
|
Effect |
ANIM_ID |
Estimate |
SE Pred |
DF |
t |
Pr >½t½ |
|
INTERCEPT |
BO241 |
– 1.836 |
1.638 |
144 |
– 1.12 |
0.264 |
|
TIME |
BO241 |
0.0563 |
0.0417 |
144 |
1.35 |
0.179 |
|
INTERCEPT |
BO322 |
– 4.014 |
1.638 |
144 |
– 2.45 |
0.0154 |
|
TIME |
BO322 |
0.084 |
0.0417 |
144 |
2.03 |
0.044 |
|
INTERCEPT |
BO326 |
– 3.53 |
1.638 |
144 |
– 2.16 |
0.032 |
|
TIME |
BO326 |
0.0676 |
0.0417 |
144 |
1.62 |
0.107 |
|
INTERCEPT |
BO209 |
4.698 |
1.838 |
144 |
2.87 |
0.0047 |
|
TIME |
BO209 |
– 0.095 |
0.0417 |
144 |
– 2.28 |
0.024 |
|
INTERCEPT |
BOL37 |
0.8398 |
1.838 |
144 |
0.51 |
0.608 |
|
TIME |
BO37 |
– 0.0359 |
0.0417 |
144 |
– 0.86 |
0.39 |
|
INTERCEPT |
BO1 |
8.841 |
1.898 |
144 |
2.35 |
0.020 |
|
TIME |
BO1 |
– 0.0777 |
0.0417 |
144 |
– 1.86 |
0.064 |
|
INTERCEPT |
ND60 |
– 2.1658 |
1.638 |
144 |
– 1.32 |
0.188 |
|
TIME |
ND60 |
0.0338 |
0.0417 |
144 |
0.81 |
0.418 |
|
INTERCEPT |
ND66 |
3.3628 |
1.638 |
144 |
2.05 |
0.041 |
|
TIME |
ND66 |
– 0.0512 |
0.0417 |
194 |
– 1.23 |
0.222 |
|
INTERCEPT |
ND72 |
– 2.688 |
1.638 |
144 |
– 1.64 |
0.102 |
|
TIME |
ND72 |
0.057 |
0.0417 |
144 |
1.37 |
0.174 |
|
INTERCEPT |
ND73 |
0.2487 |
1.638 |
144 |
0.15 |
0.879 |
|
TIME |
ND73 |
– 0.0248 |
0.0417 |
144 |
– 0.60 |
0.552 |
|
INTERCEPT |
ND74 |
– 2.8312 |
1.638 |
144 |
– 1.73 |
0.086 |
|
TIME |
ND74 |
0.0600 |
0.0417 |
144 |
1.44 |
0.152 |
|
INTERCEPT |
ND75 |
4.073 |
1.638 |
144 |
2.49 |
0.014 |
|
TIME |
ND75 |
– 0.0749 |
0.0417 |
144 |
– 1.80 |
0.0747 |
For instance, for the first animal, BO241, the fitted relationship is
BO241 : PCV =34.22–1.836+(–0.413+0.0563) t
=32.38–0.357 t
These regression lines can be determined for each animal and are presented in the following figure.
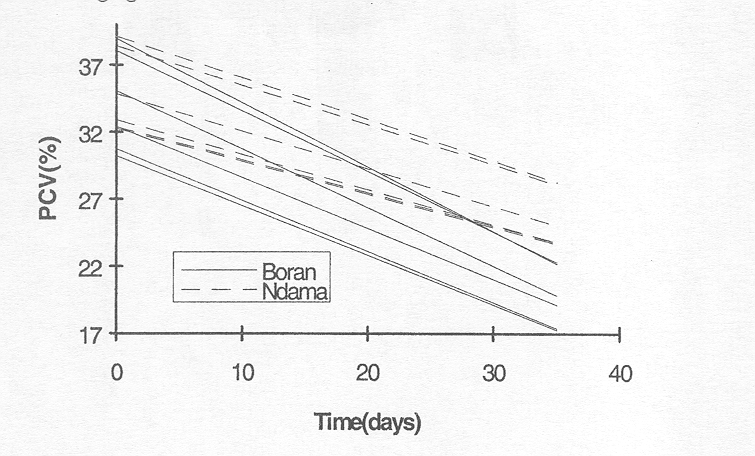
Figure 3.4.2 Change of PCV in time for individual animals based on Model 3. Bold dots and dashes indicate more than one animal with this line.
Compared with the previous analysis this figure demonstrates that PCVs tend to decrease more rapidly the higher the initial PCV. This illustrates the negative covariance between slope and intercept observed in the fitting of the model.